René Ketting
Biology of Non-Coding RNA

The major focus of my lab is gene regulation by small RNAs. Small RNAs are regulatory non-coding RNAs that have significant effects on translation, RNA stability and on chromatin. We use transgenics, genetics and biochemical and structural approaches to unravel the molecular pathways that are steered by small RNAs, focusing on a cell type that is rejuvenated every generation: germ cells.
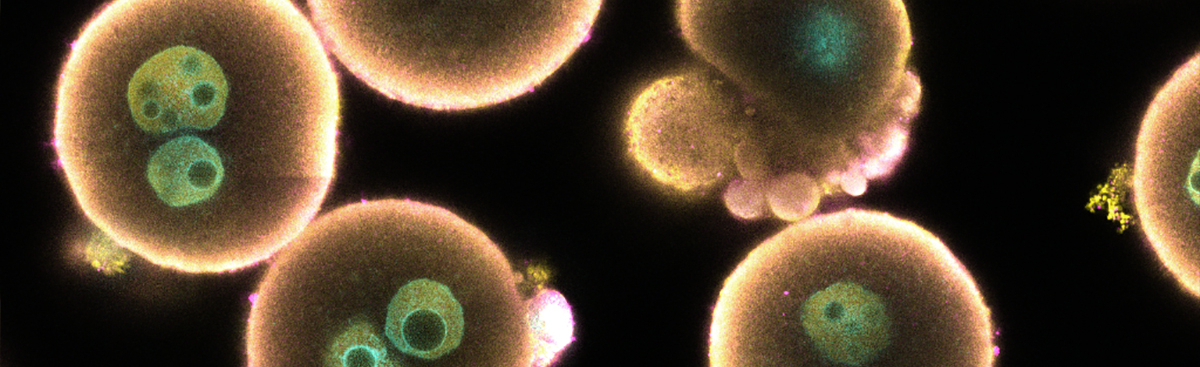
One of the major questions we address is how certain transcripts are selected to be turned into small regulatory RNAs, while others are not. This is crucial to understand, as these molecules fully determine the target specificity of the regulatory processes. In addition, we work to understand how small RNA pathways connect to other aspects of the germ cell's gene regulatory programmes in order to understand how this cell type can avoid mortality. Finally, an important current aim is to understand how phase separation acts to compartmentalise different steps in small RNA-mediated gene regulation. This is relevant in light of the many age-related diseases in which protein-RNA aggregates form and disturb cellular function. Germ cells can teach us how physiologically functional aggregates can be formed, controlled and used to control gene activity.
Positions held
- Since 2012: Scientific Director, IMB, Mainz; Professor, Faculty of Biology, Johannes Gutenberg University (JGU), Mainz
- 2010 - 2013: Professor of Epigenetics in Development, University of Utrecht
- 2005 - 2012: Group Leader, Hubrecht Institute, Utrecht
- 2000 - 2004: Postdoctoral Researcher, Hubrecht Laboratory, Utrecht
- 2000: Postdoctoral Researcher, Cold Spring Harbor Laboratories
Education
- 2000: PhD in Molecular Biology, Netherlands Cancer Institute, Amsterdam
- 1994: Masters in Chemistry, University of Leiden
Selected publications by René Ketting
Bronkhorst AW#, Lee CY, Möckel MM, Ruegenberg S, de Jesus Domingues AM, Sadouki S, Piccinno R, Sumiyoshi T, Siomi MC, Stelzl L, Luck K# and Ketting RF# (2023) An extended Tudor domain within Vreteno interconnects Gtsf1L and Ago3 for piRNA biogenesis in Bombyx mori. EMBO J, 42:e114072 (#indicates joint correspondence) Link
Podvalnaya N*, Bronkhorst AW*, Lichtenberger R, Hellmann S, Nischwitz E, Falk T, Karaulanov E, Butter F, Falk S# and Ketting RF# (2023) piRNA processing by a trimeric Schlafen-domain nuclease. Nature, 622:402–409 (*indicates joint contribution, #indicates joint correspondence) Link
Schreier J, Dietz S, Boermel M, Oorschot V, Seistrup AS, de Jesus Domingues AM, Bronkhorst AW, Nguyen DAH, Phillis S, Gleason EJ, L’Hernault SW, Phillips CM, Butter F and Ketting RF (2022) Membrane-associated cytoplasmic granules carrying the Argonaute protein WAGO-3 enable paternal epigenetic inheritance in Caenorhabditis elegans. Nat Cell Biol, 24:217–229 Link
Perez-Borrajero C, Podvalnaya N*, Holleis K*, Lichtenberger R, Karaulanov E, Simon B, Basquin J, Hennig J#, Ketting RF# and Falk S# (2021) Structural basis of PETISCO complex assembly during piRNA biogenesis in C. elegans. Genes Dev, 35:1304–1323 (*indicates joint contribution, #indicates joint correspondence) Link
Roovers EF, Kaaij LJT, Redl S, Bronkhorst AW, Wiebrands K, de Jesus Domingues AM, Huang HY, Han CT, Riemer S, Dosch R, Salvenmoser W, Grün D, Butter F, van Oudenaarden A and Ketting RF (2018) Tdrd6a regulates the aggregation of Buc into functional subcellular compartments that drive germ cell specification. Dev Cell, 46:285–301.e9 Link